Figures
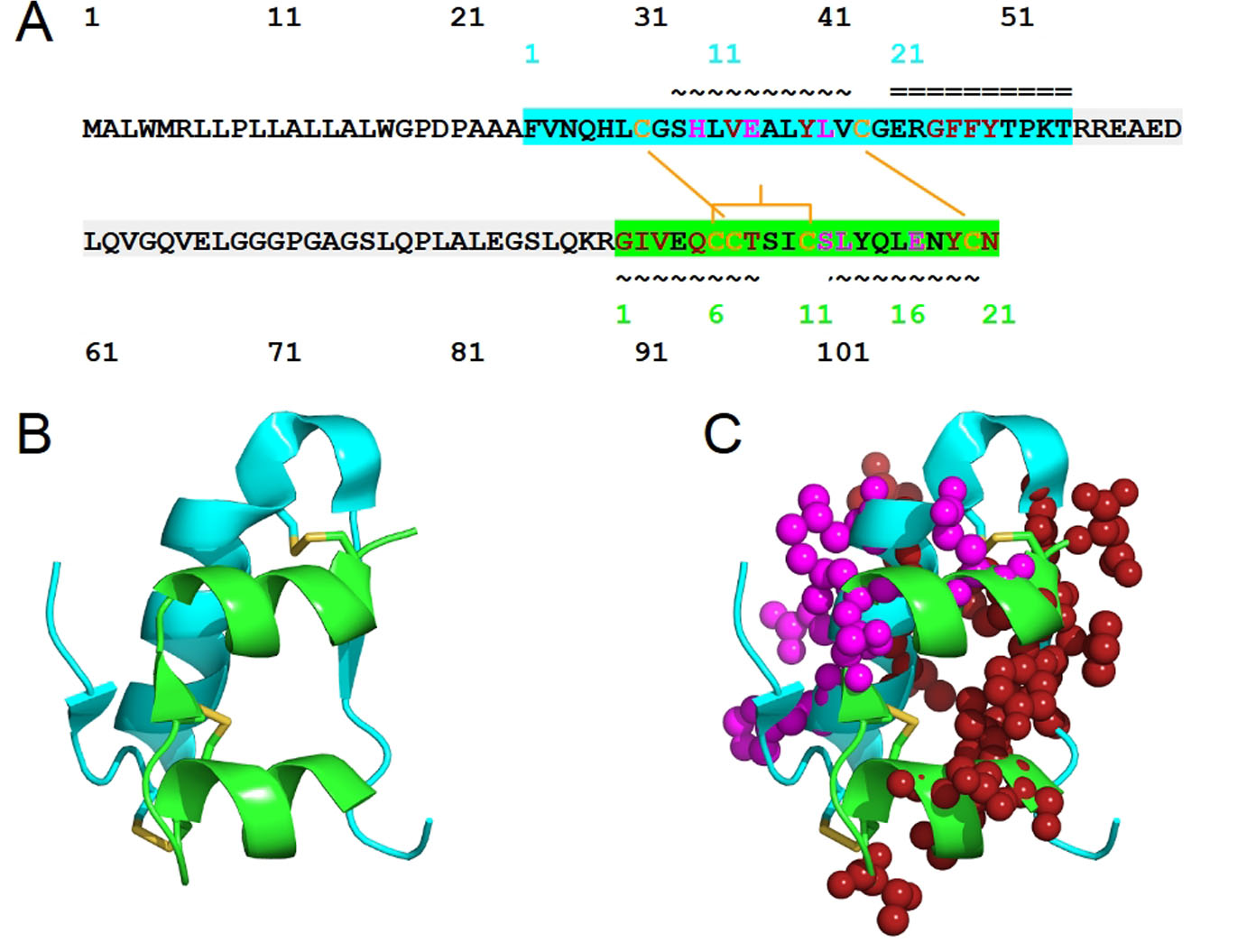
Figure 1. The structure of human insulin. (A) The sequence of human insulin in the form of preproinsulin (UniProt ID P01308 numbering in black) is highlighted according to the respective chains of insulin: chain B (cyan) and chain A (green). C-peptide is highlighted in light gray. Cysteine residues involved in inter-chain disulfide bridges (Cys7 of chain A and Cys7 of chain B, Cys20 of chain A and Cys19 of chain B) and those that contribute to intra-chain A disulfide bridge, Cys6 and Cys 11, are shown in yellow-orange. The secondary structure of human insulin consists of three alpha-helices formed by chain A residues Gly1-Thr8, Ser12-Asn18 and chain B residues Ser9-Cys19. The residue numbers of insulin chains B and A are shown in cyan and green, respectively. The symbols ~ and = indicate α-helices and β-strands, respectively. Insulin residues that bind to IR sites 1 and 2 are shown in red and magenta, respectively. (B) The structure of human insulin with the same coloring scheme as (A) and the insulin residues that bind to IR sites 1 and 2 are shown in spheres (C).
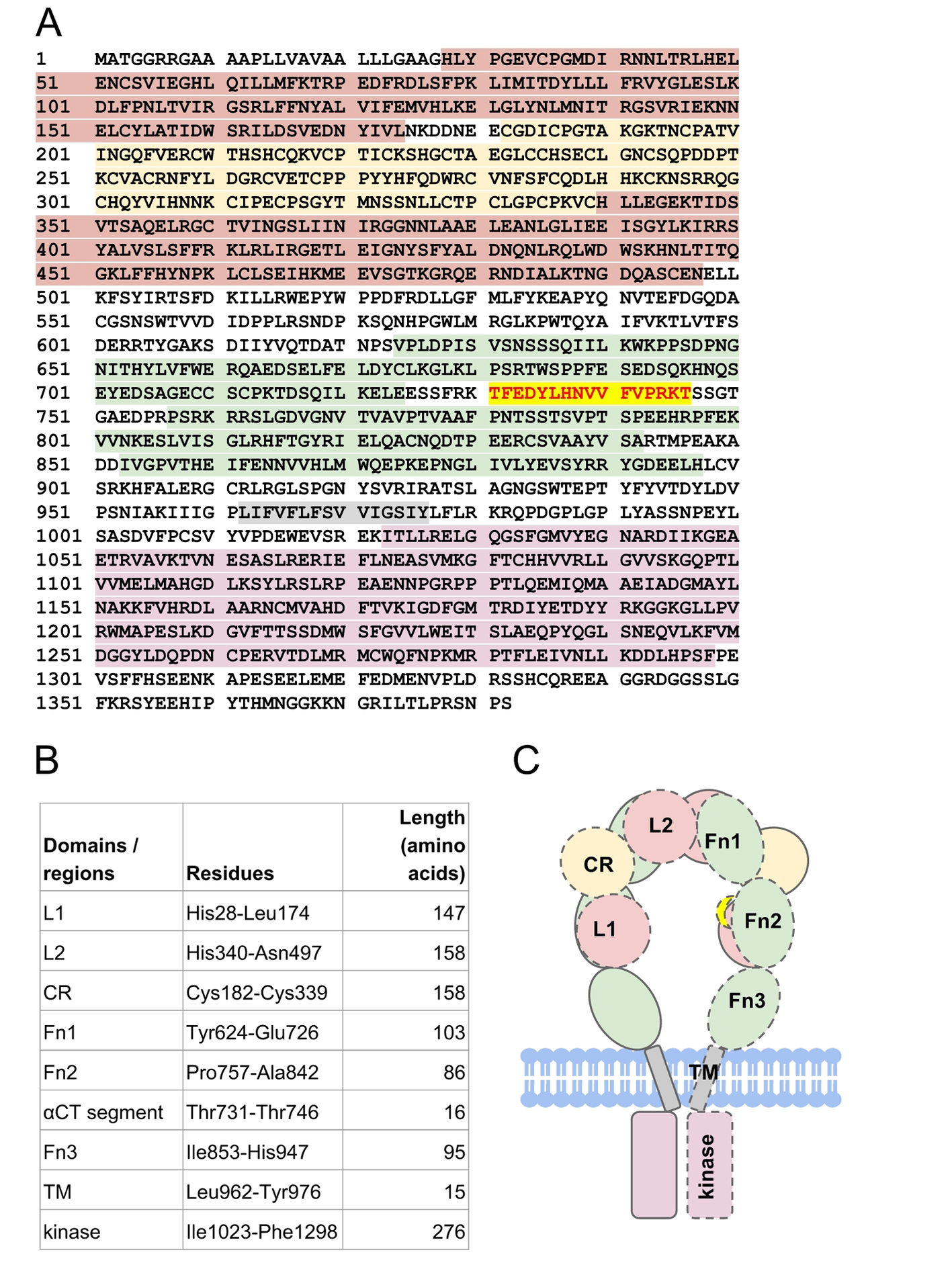
Figure 2. The structure of human insulin receptor. (A) The sequence of human insulin receptor (UniProt ID P06213) is highlighted according to its respective domains (B): leucine-rich domains (red maroon), cysteine-rich domain (yellow), type III fibronectin domains (green), transmembrane (gray) domain, and tyrosine kinase domain (pink). Residues Thr731-Thr746 (red scarlet) forms the αCT segment that is important for insulin binding. (C) Schematic representation of insulin receptor domains in the absence of insulin. The domains of a half-receptor and the other half-receptor of insulin receptor are distinguished by solid and dashed lines.
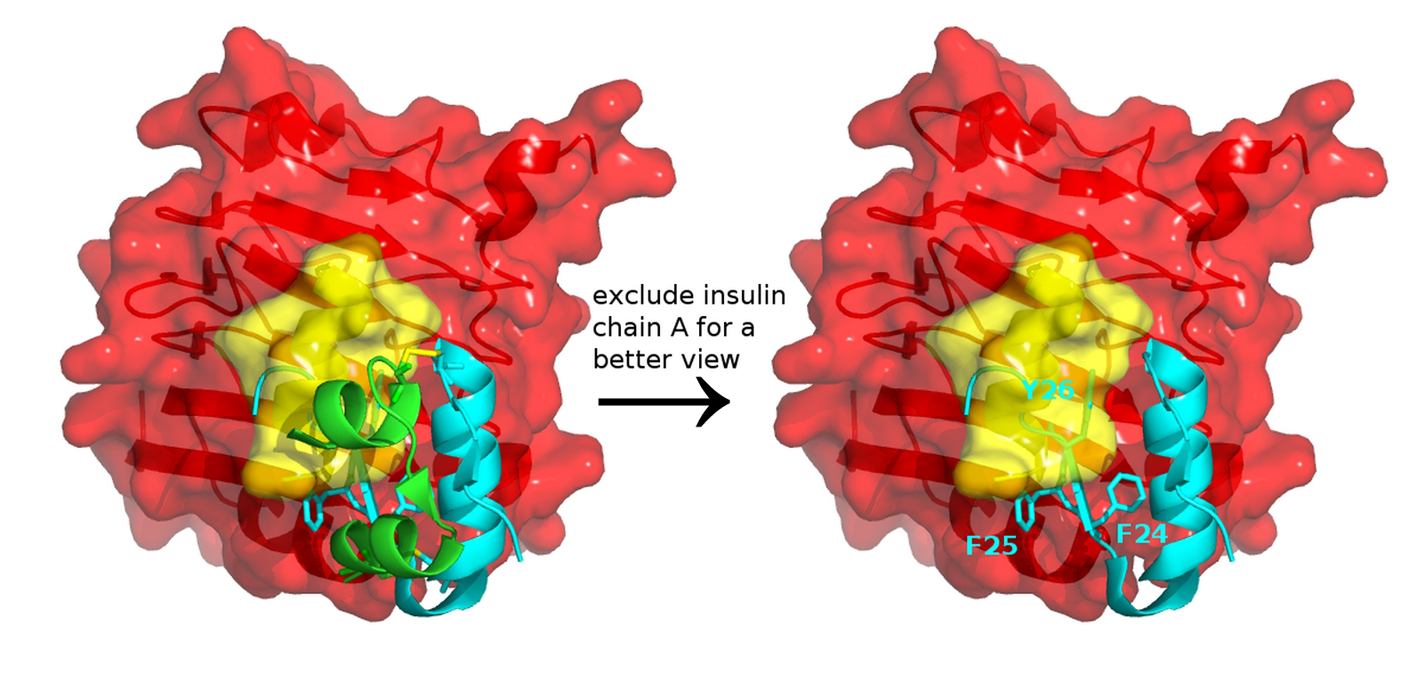
Figure 3. Insulin and insulin receptor interaction. Docking a full-length insulin (PDB entry 4EY1) to the IR structure (PDB entry 3W14) containing αCT (yellow) and L1 domain (red) highlights that conformational change and structural dynamics have to take place to avoid a steric clash between the aromatic residues of insulin and the αCT segment of IR.
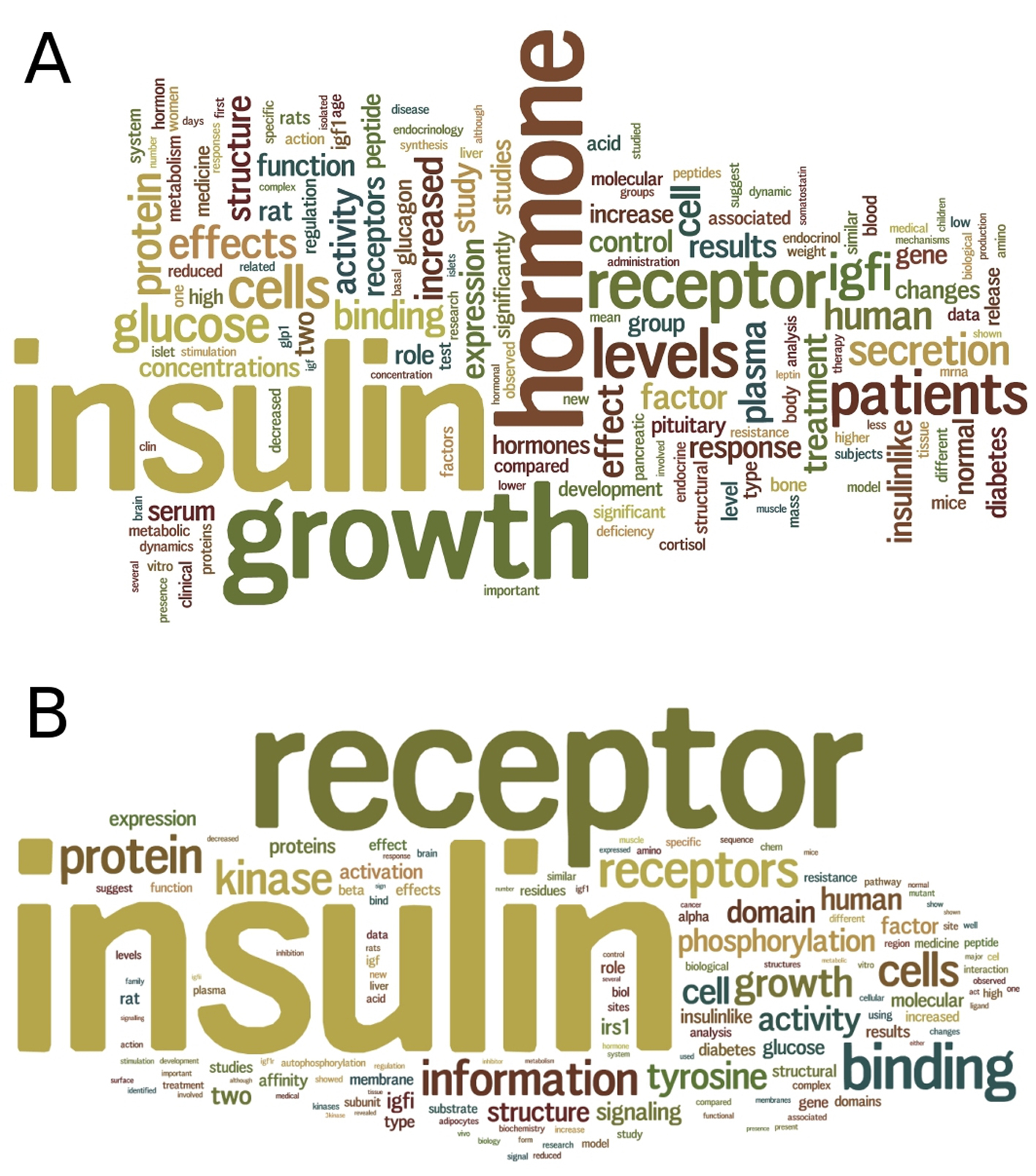
Figure 4. Text mining of insulin and insulin receptor literature. (A) A summary wordcloud generated from text mining of 2,901 abstracts of articles in PubMed for insulin hormone, provides a clue on the relationships of insulin with other hormones including glucagon, leptin and cortisol. (B) A summary wordcloud generated from text mining of 2,201 abstracts of articles in PubMed for insulin receptor, provides a clue on the binding of not only insulin but also insulin growth factor (IGF) to the insulin receptor.

Figure 5. Sequence similarity among insulin and IGFs. (A) Pairwise sequence identity (%). (B) Multiple sequence alignment performed using Clustal Omega shows two relatively conserved regions (boxed) spanning residues L30-Y50 and residues G90-C109 of insulin hormone, which correspond to the cleaved chains B and A, respectively.
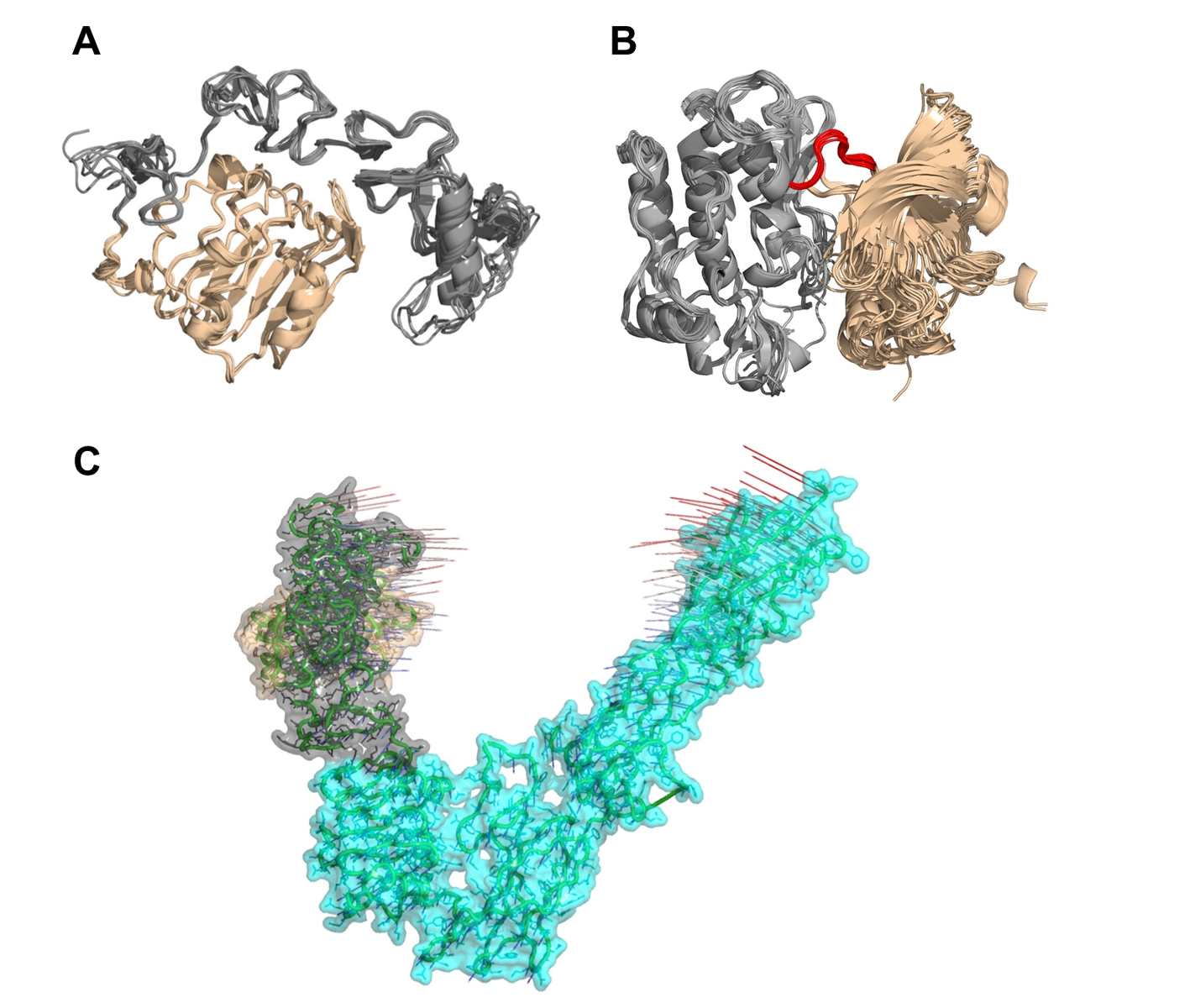
Figure 6. Multiple structural alignments of insulin receptors (IR) to identify the clustroid structure. (A) The experimentally available structures of the L1 domain (wheat) and the cysteine-rich domain (gray) of IR show subtle structural dynamics. (B) It appears that the N-terminal region (wheat) of IR residues Asp1014-Lys1310 undergoes rotation at loop Met1078-Cys1083 (red) with respect to the C-terminal region (gray), resulting in structural dynamics. (C) Normal mode analysis on PDB entry 3LOH_E suggests that the N-terminal L1 domain (wheat) and the CR domain (gray) show a potential for large-scale motions towards each other and the C-terminal of IR ectodomain.





