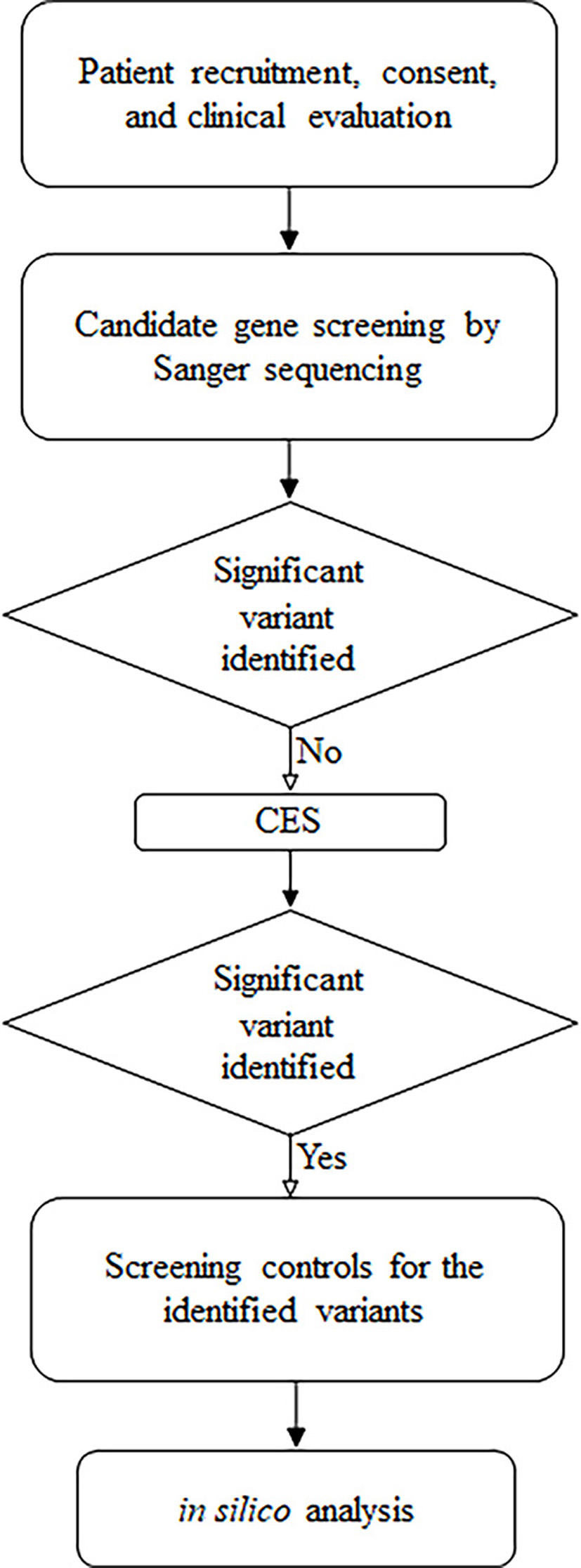
Figure 1. Summary of the followed research design.
| Journal of Endocrinology and Metabolism, ISSN 1923-2861 print, 1923-287X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Endocrinol Metab and Elmer Press Inc |
| Journal website https://www.jofem.org |
Short Communication
Volume 12, Number 6, December 2022, pages 188-197
Identification of Two Rare Homozygote Missense Variants in the IRS1 Gene in a Patient With Early Gestational Diabetes: A Case Study
Figures

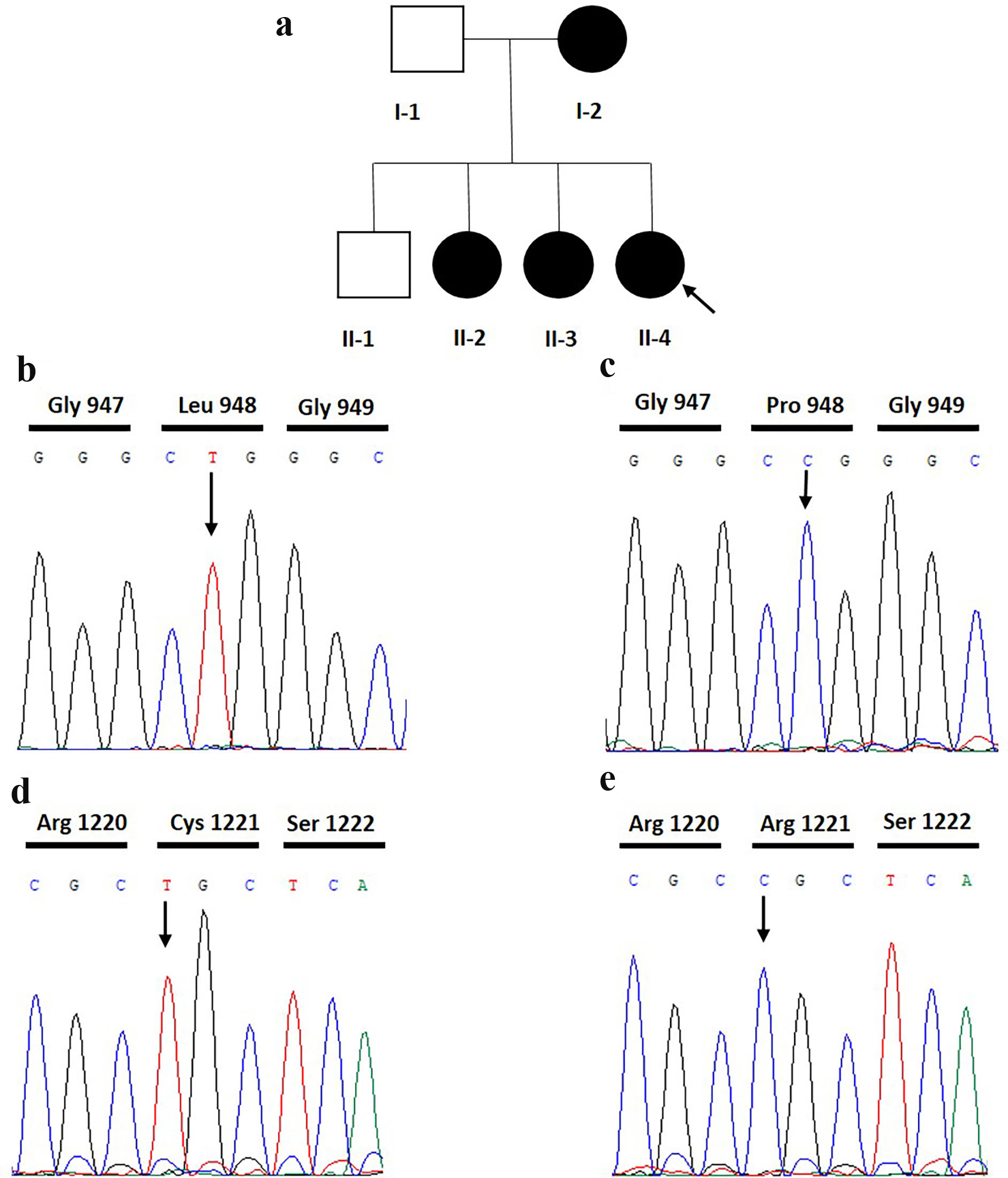
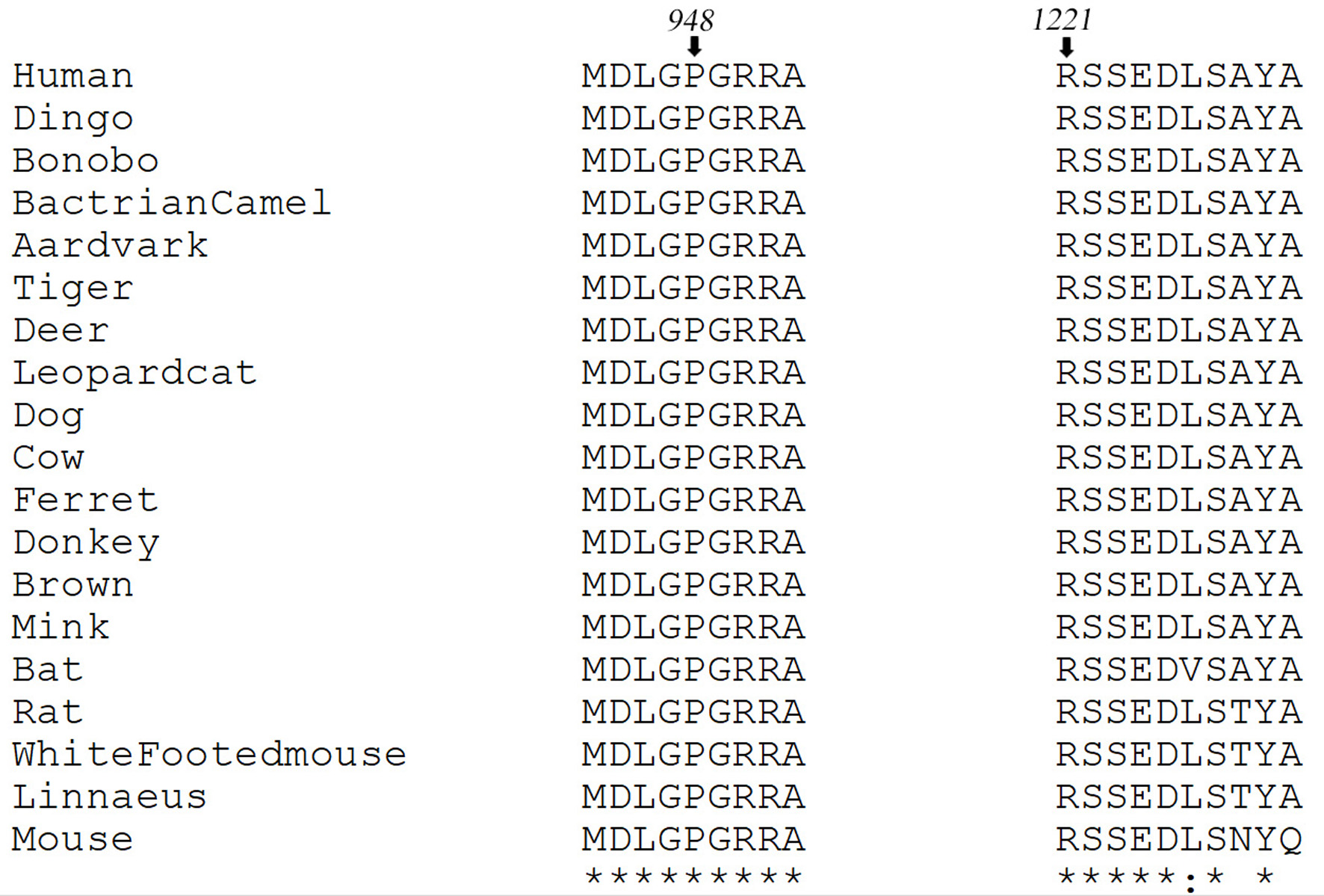
Tables
| RefSNP number | Position in genomic DNA | Position in CDS | Position in protein | Global MAF | ClinVard | References |
|---|---|---|---|---|---|---|
| aMAF of the global population from 1000 Genomes Study. bMAF of the global population from gnomAD. cThe positions are in the transcript ENST00000400024.6. dClinVar interpretation and clinical significance for the phenotype of maturity-onset diabetes of the young type 3. CDS: coding DNA sequence; MAF: minor allele frequency. | ||||||
| rs1169289 | Chr12:g.121416622C>G | c.51C>G | p.Leu17= | G = 0.4285a G = 0.4645b | Benign | [28] |
| rs2259820 | Chr12:g.121435342C>T | c.1375C>T | p.Leu459= | T = 0.3167a T = 0.3302b | Benign | [28, 29] |
| rs2259816 | Chr12:g.121435587G>T | c.1620G>Tc | p.Val540=c | T = 0.3588a T = 0.3845b | Benign | [30, 31] |
| rs1169288 | Chr12:g.121416650A>C | c.79A>C | p.Ile27Leu | C = 0.2985a C = 0.3479b | Benign | [32-34] |
| rs2464196 | Chr12:g.121435427G>A | c.1460G>A | p.Ser487Asn | A = 0.3177a A = 0.3296b | Benign | [28, 35] |
| rs2464195 | Chr12:g.121435475G>A | c.1508G>Ac | p.Arg503Hisc | A = 0.3596a A = 0.3800b | Benign | [36] |
| rs370373307 G>A MAF = 0.00007569 | |||
|---|---|---|---|
| G | A | ||
| The estimated probability that these two variants occur together among the same individual is equal to 0%. Adapted from gnomAD v2.1.1. MAF: minor allele frequency. | |||
| rs754239320 4G>A MAF = 0.00005627 | G | 248,813 | 19 |
| A | 14 | 0 | |