| Journal of Endocrinology and Metabolism, ISSN 1923-2861 print, 1923-287X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Endocrinol Metab and Elmer Press Inc |
| Journal website https://www.jofem.org |
Short Communication
Volume 12, Number 6, December 2022, pages 188-197
Identification of Two Rare Homozygote Missense Variants in the IRS1 Gene in a Patient With Early Gestational Diabetes: A Case Study
Maab Babiker Mansoura, Uzma Inayata, Aml Mohamed Nadab, c, Hassen Hadj Kacema, d
aDepartment of Applied Biology, College of Sciences, University of Sharjah, Sharjah, United Arab Emirates
bThe Faculty of Medicine, Mansoura University, Egypt
cSaudi German Hospital, Sharjah, United Arab Emirates
dCorresponding Author: Hassen Hadj Kacem, Department of Applied Biology, College of Sciences, University of Sharjah, Sharjah, UAE
Manuscript submitted October 24, 2022, accepted December 13, 2022, published online December 30, 2022
Short title: IRS1 Gene in Early Gestational Diabetes
doi: https://doi.org/10.14740/jem842
| Abstract | ▴Top |
Background: Gestational diabetes mellitus (GDM) is one of the most common pregnancy complications and has a rising prevalence worldwide. Environmental and genetic factors contribute to GDM risk. Describing familial cases of GDM can help identify the disease’s genetic components.
Methods: Here, we report the case of an Emirati female patient affected with early GDM and familial history of diabetes with a significant level of insulin resistance. As a first step, we investigated the GCK and HNF1A gene sequences by direct sequencing and then performed a clinical exome sequencing (CES) of the patient’s genomic DNA covering 6,670 genes.
Results: Our findings showed the absence of any pathogenic variants in the GCK and HNF1A gene sequences. In addition, the CES excluded all maturity-onset diabetes of the young (MODY)-related genes. However, two rare homozygous variants in the insulin receptor substrate 1 (IRS1) gene were identified: p.Pro948Leu (gnomAD minor allele frequency (MAF) = 7.5 × 10-5) and p.Arg1221Cys (gnomAD MAF = 5.6 × 10-5). These variants were absent in a set of healthy Emirati individuals. Both variants are highly conserved among mammalians but have never previously been reported among the same haplotype or individual. p.Pro948Leu and p.Arg1221Cys are localized close to two important functional phosphorylation sites recognized by PI3K (hTyr941) and SHP2 (hTyr1229), respectively. Moreover, the independent and combined effect of the two variants on protein stability was predicted to be destabilizing.
Conclusion: Our investigation emphasized the role of downstream regulators of insulin signaling in GDM pathophysiology and identified IRS1 as a candidate gene to explain chronic insulin resistance. In addition, the patient showed significant health improvement after lifestyle modifications and oral antidiabetic administration, even after withdrawing insulin injections.
Keywords: Gestational diabetes mellitus; Clinical exome sequencing; Sanger sequencing; IRS1 gene; Insulin resistance
| Introduction | ▴Top |
Gestational diabetes mellitus (GDM) is defined as any degree of glucose intolerance first diagnosed during pregnancy. Many, if not most, cases of GDM represent preexisting diabetes that is only discovered during pregnancy [1]. With the increasing prevalence of obesity and diabetes, more type 2 diabetes mellitus (T2DM) is diagnosed in women of reproductive age and early pregnancy [2]. So, it is reasonable to test women with risk factors for T2DM early in pregnancy and, better yet, before conception [3, 4].
The global estimate of hyperglycemia in pregnancy in 2021 was around 21.1 million (16.7%) live birth cases. Around 80.3% of these hyperglycemic cases were due to GDM, while 10.6% were because of pre-pregnancy diabetes and 9.1% were the results of other types of diabetes detected during the pregnancy. GDM affects one in eight births worldwide but mostly in low- and middle-income countries where maternal care access is limited [5].
During a normal healthy pregnancy, the maternal body goes through major metabolic changes to support the fetus’s growth. During early gestation, from week 11 to week 16, insulin sensitivity increases, promoting glucose uptake and storage for the upcoming increase in energy demand during later gestation. As pregnancy progresses, hepatic gluconeogenesis increases by 30%, and glucose sensitivity decreases by 50-60% [6]. The reason for the elevated gestational glucose is that it is the main source of fuel for the fetus and the placenta. Hence, placental glucose transport is mediated by glucose transporter 1 (GLUT1) via passive diffusion. To maintain euglycemia, β cells undergo hyperplasia and hypertrophy and increase insulin secretion by 2-3 fold [7, 8]. Most (about 80%) GDM cases present as β-cell dysfunction on a background of chronic insulin resistance, to which insulin resistance of pregnancy is added [9]. Therefore, overlapping etiologies between the different forms of diabetes generated much debate, and it is not excluded [9, 10].
GDM is a heterogeneous disorder influenced by various environmental, genetic, and epigenetic factors. Multiple observations point to the genetic predisposition of GDM, including clustering of GDM within families with a history of diabetes, the higher recurrence rate in subsequent pregnancies, and risk rate disparency among different ethnic groups [11]. Based on the suggestion that T2DM and GDM share a common genetic background, many genetic studies attempted to investigate variants and genes that confer a high risk of developing T2DM with GDM. The largest group of loci are in genes related to insulin secretion, such as TCF7L2, GCK, KCNJ11, CDKAL1, IGF2BP2, and MTNR1B, all of which are also associated with T2DM [12]. Other genes studied concerning GDM have been categorized into insulin and insulin signaling genes, lipid and glucose metabolism genes, maturity-onset diabetes of the young (MODY) genes, and other genes [13].
In the present study, an Emirati woman with diabetes mellitus was diagnosed during her first pregnancy at the age of about 23 years. She had a similar history of diabetes development in her mother and sisters. The patient was examined genetically using direct and clinical exome sequencing (CES). Two rare variants in the insulin receptor substrate 1 (IRS1) genes were identified (c.2843C>T, p.Pro948Leu, g.227660612G>A, rs370373307 and c.3661C>T, p.Arg1221Cys, g.227659794G>A, rs754239320).
| Materials and Methods | ▴Top |
This was a case study carried out by the Department of Applied Biology (University of Sharjah) in association with the Saudi German Hospital. The consenting recruited patient was clinically evaluated, and the molecular screening was done initially by candidate genes mutational sequencing using Sanger sequencing. Since none of the identified variants showed clinical significance, CES was performed. After filtration and analysis, control subjects were screened for the CES-identified variants, followed by in silico analysis (Fig. 1). All experimental procedures used in this study were approved by the University of Sharjah Research Ethics Committee (No. REC-15-11-P004) and performed in accordance with the relevant guidelines and regulations.
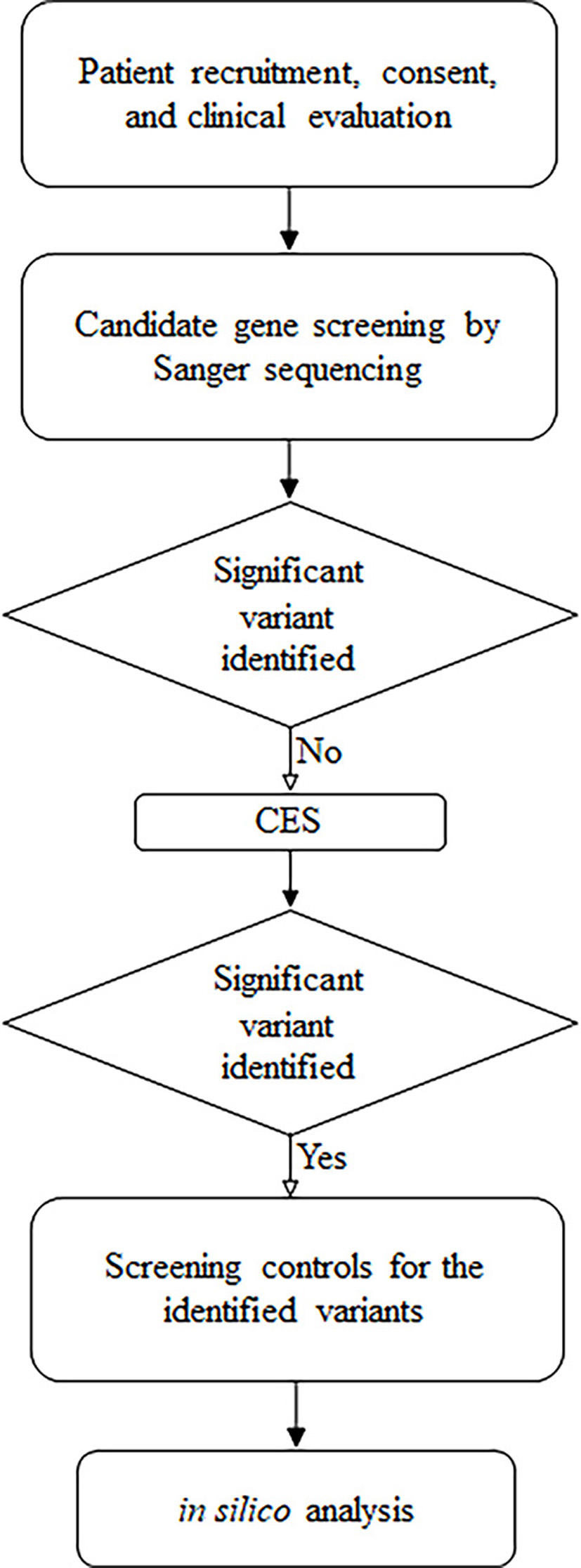 Click for large image | Figure 1. Summary of the followed research design. |
Patients
Our patient is a 33-year-old female. She has had diabetes mellitus for 15 years. She was diagnosed with diabetes early, at 6 weeks of gestation in her first pregnancy, at the age of 18 years. This was discovered during early pregnancy medical checkups with no symptoms referring to diabetes. Diabetes was controlled by diet and physical activity together with metformin. The patient became pregnant again 1 year after the delivery of her first baby with the same management of diabetes. According to her, her blood glucose usually returned to normal during a postpartum hospital stay for a few days, but she never checked after that, whether after the first or second delivery. Her third pregnancy was 3 years after her second pregnancy, during which she needed insulin and metformin for glycemic control. Since her third pregnancy, she has had persistent diabetes and has been using insulin analogues at different regimens and types. She did not benefit from sulphonylurea gliclazide. She reports better control when using a sitagliptin-metformin combination in a dose of 50/1,000 mg twice daily, but she never stopped insulin. In her fourth pregnancy, her blood glucose level was controlled by a healthy lifestyle and metformin, but insulin was added in the third trimester. Since her fourth delivery, she has been using insulin analogues, both long- and short-acting (basal-bolus), with at least three injections per day without oral antidiabetic medication.
This patient was never compliant with diet or exercise except during her pregnancy periods; otherwise, she has a completely sedentary lifestyle with heavy carbohydrates and fast-food meals together with sugar-laden desserts. Clinically she has a body mass index (BMI) of 32.3 kg/m2 and a wide waist-hip ratio of 0.95 (as per WHO definition). She had goiter, a blood pressure of 140/100 mm Hg, and a heart rate of 103 bpm. She does not have acanthosis or pseudoacanthosis nigricans. Funduscopy revealed early non-proliferative diabetic retinopathy (NPDR). In addition, serological investigations reveal that our patient is negative for glutamic acid decarboxylase antibodies (< 5 IU/mL (Ref. < 10)), islet cell antibodies, and insulin antibodies (1.32 U/mL (Ref. < 12)). On the other hand, insulin resistance was assessed for the patient by calculating the Homeostatic Model Assessment of Insulin Resistance (HOMA-IR) value using the patient’s first biochemical investigation (fasting insulin, 35.47 pmol/L; fasting plasma glucose, 308.01 mg/day). The HOMA-IR value of 4.5 showed significant insulin resistance.
This patient has a similar family history in her mother and two sisters, who were diagnosed with diabetes in their first pregnancy. Neither her father nor her brother has been diagnosed with diabetes (Fig. 2a).
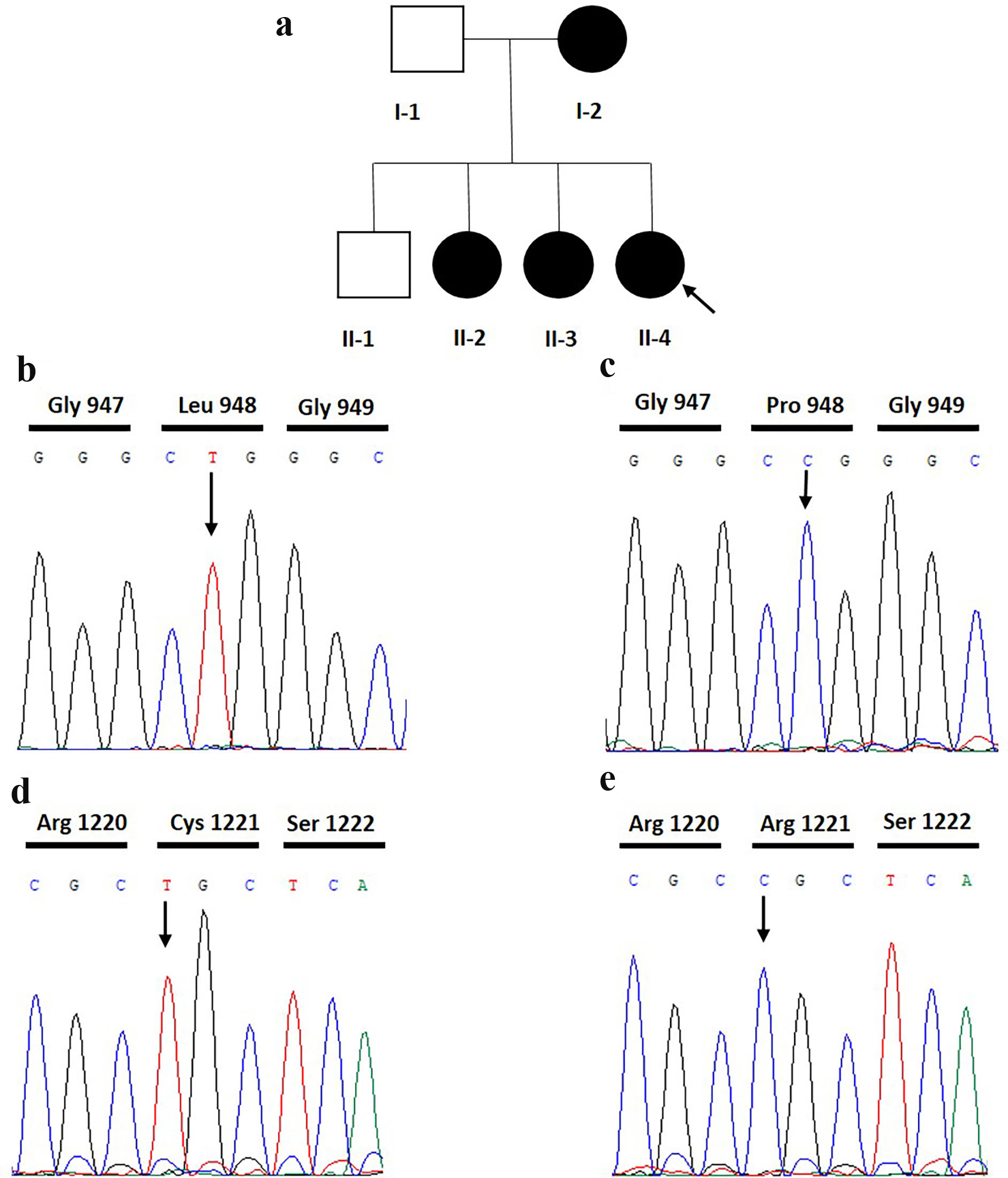 Click for large image | Figure 2. Pedigree and electropherograms of the index patient. (a) Pedigree of the family of the studied patient with GDM. Arrow is the index patient. (b) Electropherograms of the homozygous mutant index patient with c.2843C>T variant. (c) The IRS1 homozygous wild type individual missing the c.2843C>T variant. (d) The homozygous mutant index patient with c.3661C>T variant. (e) IRS1 homozygous wild type individual missing the c.3661C>T variant. Arrow is the position of the variants. |
Genetic screening
After informed consent was obtained, a blood sample was collected from the patient’s peripheral veins. Genomic DNA was extracted from the blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Germany). The extracted DNA was quantified using NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Scientific™, Massachusetts, USA).
The coding exons and their flanking intron-exon regions of GCK (NG_008847.2) and HNF1A (NG_011731.2) genes were screened for mutations by Sanger sequencing. First, specific primers were designed using Primer3web version 4.1.0 [14] (Supplementary Material 1, www.jofem.org). Polymerase chain reactions (PCRs) were performed using Promega’s PCR Master Mix, 2 × (Promega Corporation, Wisconsin, USA). The final PCR reaction volume of 50 µL contained 50 ng of genomic DNA. The PCR conditions were as follows: initial denaturation at 95 °C for 3 min; 40 cycles of 94 °C for 30 s, 54 - 61 °C for 30 s (depending on each primer pair’s optimization), and 72 °C for 45 s; and final extension at 72 °C for 5 min. The amplified amplicons were purified using either ExoSAP-IT™ PCR Product Cleanup Reagent (Thermo Scientific™, Massachusetts, USA) or QIAquick Gel Extraction Kit (Qiagen, Germany). Then, the sequencing was performed using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystem, Thermo Scientific™, Massachusetts, USA). As recommended by the protocol, the ethanol/EDTA/sodium acetate precipitation method was used to purify the sequencing reaction products before running the capillary analysis in Applied Biosystem’s Genetic Analyzer 3500 (Thermo Scientific™, Massachusetts, USA). The resulting sequences were aligned with the human reference sequences of GCK and HNF1A by the Basic Local Alignment Search Tool (BLAST).
CES
CES was performed on the patient’s DNA using MedGenome Clinical Exome V4 and Illumina technology covering 6,670 clinically related genes (about 30 Mb) with known associations to inherited diseases. The protocol was conducted according to the manufacturer’s instructions. In brief, the library preparation is done by genomic DNA fragmentation and adding adaptors containing sequencing binding sites, indexes, and regions complementary to the flow cell’s oligos. Then, the library was enriched by hybridizing biotinylated probes to targeted regions and sequenced on HiSeq 2000 sequencing system (Illumina Inc., California, USA). Sequences were aligned to the genome version hg19/GRCh37 using BWA-MEM. Realignment and recalibration were performed using GATK-lite (V2.3-9).
The annotated variants from the CES data were filtered considering the following parameters: new variants and variants with frequencies lower than 0.01 in ExAC Browser, 1000 Genomes Project, and dbSNP retained. All variants with coverage greater than 20 were analyzed. A special focus was given to a list of 133 genes related to diabetes, glucose metabolism, and the insulin pathway [15]. In addition, the functional impacts of the selected coding variants were predicted using the following tools: Sorting Intolerant From Tolerant (SIFT) [16], Protein Variation Effect Analyzer (PROVEAN) [17], and Polymorphism Phenotyping v2 (PolyPhen-2) [18].
The selected variants were confirmed by direct sequencing by the following primers: Pro948Leu-F: 5′ TTGGGAGTGATCAGTCTGGC 3′, p.Pro948Leu-R: 5′ CTGACGGGGACAACTCATCT 3′, Arg1221Cys-F: 5′ GGGGTTTGGAGAATGGTCTT 3′, and Arg1221Cys-R: 5′ GCAGAGGCGAAGAACAGAAT 3′. In addition, 42 unrelated non-diabetic healthy subjects from the UAE population were screened for candidate mutations.
Sequence analysis and silico predictions
Multiple sequence alignment
The human IRS1 protein sequence was aligned with orthologous mammalian sequences to evaluate the conservation of mutated residues. Sequences were retrieved through the NCBI database, and multiple sequence alignment was performed using the Clustal Omega program available at the EBI server.
Protein modelling: model of the full-length IRS1
As the full-length structure of IRS1 is not experimentally determined, the FASTA sequence of human IRS1 (UniProt ID: P35568) was downloaded from the UniProt database [19]. The structure model of wild type human IRS1 was predicted through the I-TASSER server using the threading method [20, 21]. Based on the scoring functions, the best model was selected. A scoring function (C-score) based on the relative clustering structural density and the consensus significance score of multiple threading templates was used to estimate the accuracy of the I-TASSER predictions. The stereochemical quality of the selected model generated was assessed using QMEAN [22, 23] and ProSA webserver [24, 25].
Effect of mutation on protein stability
The predicted wild type protein structure was used to see the effect of the two missense mutations found in the clinical study p.Pro948Leu and p.Arg1221Cys on protein stability using PrempPS [26] and MAESTRO web servers [27]. PremPS evaluates the effects of single mutations on protein stability by calculating the quantitative changes in unfolding Gibbs free energy, while MAESTRO provides predicted ΔΔG values along with a corresponding prediction quality measure for multiple mutations. The predictions are based on the protein structure for both servers.
| Results | ▴Top |
Direct Sanger sequencing was performed on the 10 exonic sequences of the GCK and HNF1A genes. No variants were found in the GCK gene. On the other hand, screening the HNF1A gene sequences revealed six variants: three missenses and three synonymous variants (Table 1) [28-36].
 Click to view | Table 1. Description of HNF1A Variants Detected in the Index Patient |
The three homozygous synonymous variants are p.Leu17=, p.Leu459=, and p.Val540= (ENST00000400024.6). Only one heterozygous missense variant (p.Ile27Leu, at position g.121416650A>C of exon 1) was detected. The other two missense variants are homozygous, p.Ser487Asn and p.Arg503His (ENST00000400024.6). All HNF1A variants are reported as benign by the ClinVar database.
The CES data from the patient yielded a total of 162,654 variations. These were filtered to keep only non-synonymous variants in coding regions with frequencies lower than 0.01 in dbSNP, ExAC Browser, and gnomAD and associated with diabetes, insulin pathway, and glucose metabolism. Also, the bioinformatics tools PolyPhen-2, PROVEAN, and SIFT were used to predict the pathogenicity of the remaining variants. As a result, the patient was shown to harbor two homozygous variants in the IRS1 (NG_015830.1) gene, p.Pro948Leu (c. 2843C>T, g.227660612G>A, rs370373307) and p.Arg1221Cys (c.3661C>T, g.227659794G>A, rs754239320) (Fig. 2b-e).
To validate these findings, Sanger sequencing was performed on the patient using our own designed primers spanning the regions containing the variants. We confirmed the presence of the two homozygous mutant genotypes in the patient. Furthermore, the mutant alleles of the p.Pro948Leu and p.Arg1221Cys variants were absent in 42 non-diabetic individuals. According to gnomAD (V2.1.1), both variants are very rare, with respective frequencies of 7.5 × 10-5 (19/251,030) and 5.6 × 10-5 (14/248,810). The variant p.Arg1221Cys is never found homozygous. However, the p.Pro948Leu was reported homozygous only once in a Latino female. The frequencies of p.Pro948Leu and p.Arg1221Cys among the south Asian population are 3.9 × 10-4 and 4.5 × 10-4, respectively. Although they are linked to the same gene, the two mutant alleles have never previously been described in the same individual (Table 2).
 Click to view | Table 2. The Co-Occurrence of the Two Variants p.Pro948Leu (rs370373307) and p.Arg1221Cys (rs754239320) in the Same Haplotype |
Multiple sequence alignment shows that the residues Pro948 and Arg1221 are conserved in multiple species (Fig. 3). The wild type protein structure of IRS1, as shown in Supplementary Material 2 (www.jofem.org), was selected with a C-score of -1.03 and a TM score of 0.58, which indicates a model of correct topology. The selected model was submitted to the PremPS web server, and non-covalent interactions between the mutated site and its adjacent residues in the wild type and mutant structure were predicted (Supplementary Material 2, www.jofem.org). ΔΔG (kcal/mol) predicted unfolding free energy change induced by a single mutation. The sign of the predicted score depends on the tool selected. PremPs uses positive signs to indicate destabilizing mutations and negative signs for stabilizing mutations, while MAESTRO uses opposite signs. As indicated in Supplementary Material 3 (www.jofem.org), the effect of every single variant on protein stability was predicted as ΔΔG value, which indicates that these mutations are destabilizing the protein.
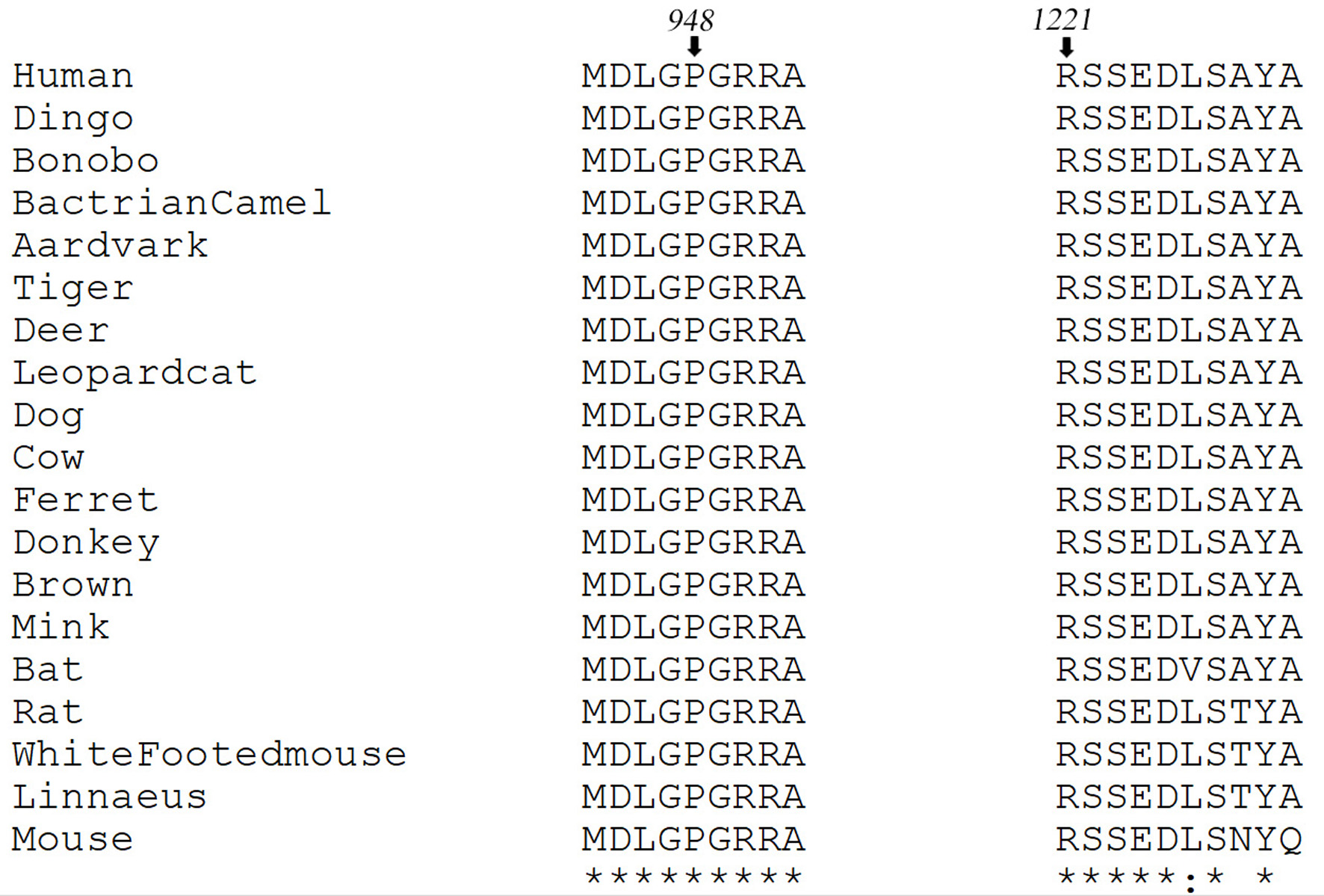 Click for large image | Figure 3. Multiple sequence alignment of human IRS1 protein sequence (NP_005535.1) with its mammalian homologs. Black arrows indicate residue positions Pro948 and Arg1221. |
MAESTRO also predicts the effect of multiple mutations on protein stability. So, the effect of p.Pro947Leu and p.Arg1221Cys mutations on IRS1 protein structure was predicted. Based on the ΔΔG value predicted by this server for these two mutations combined, these changes are considered destabilizing.
| Discussion | ▴Top |
The IRS1 gene is located on chromosome 2q36.3, consisting of two exons and 9,705 base pairs long. It encodes for the protein IRS1, which has a molecular mass of 131 kDa with an amino acid sequence of 1,242 residues. Insulin receptor (IR), a tyrosine kinase receptor, is activated upon insulin binding, which results in the autophosphorylation of IR followed by tyrosine phosphorylation of IRS1 at multiple sites generating binding sites for Src homology 2 (SH2) domain-containing proteins, such as PI3K, GRB2/SOS, and SHP2. The recruitment of PI3K to IRS1 regulates insulin-dependent processes, including glucose uptake, glycogen synthesis, and promoting hepatic gluconeogenic gene transcription [37]. IRS1 has nine YXXM motifs serving as putative PI3K binding sites at tyrosine positions: 465, 551, 612, 632, 662, 732, 941, 989, and 1012 [38, 39].
The pathogenicity of the first IRS1 variant, p.Pro948Leu was estimated to be possibly damaging (score = 0.707) by PolyPhen-2, which is in concordance with our in silico predictions to be destabilizing. This variant is nearby the PI3K binding site hTyr941, located only seven amino acids away. A study using 32P labelling and radio-sequencing of phosphopeptides proved that the SH2 domain of p58 binds to phosphorylated mTyr608 and mTyr939 in rat IRS1, corresponding to human hTyr612 and hTyr941, respectively [40]. In another study, different mutant rat IRS1 variants were constructed by site-directed mutagenesis to evaluate their interaction levels with the SH2 domain of PI3K. The construct harboring a deletion between 898 and 1146 resulted in only 40% of PI3K activity compared to the wild type [41]. In Xenopus laevis oocytes, an IRS1 mutant at four tyrosine residues (Y460F, Y608F, Y939F, and Y987F) was overexpressed. Insulin-stimulated PI3K activity and tyrosine phosphorylation levels of IRS1 were nearly undetectable, while oocyte maturation was partially decreased [42].
The most extensively studied IRS1 variant - p.Gly972Arg (rs1801278) - is located between the two tyrosine phosphorylation sites and p58 binding sites hTyr941 and hTyr989. Similarly, our variant - p.Pro948Leu - is also between the same sites. Different studies have linked the p.Gly972Arg variant with higher risks of T2DM, polycystic ovary syndrome, GDM, and osteoarthritis [43-46]. Its molecular mechanism was explained using a series of recombinant peptide fragments between IRS1 residues 910 to 1027 containing the polymorphism. The presence of the polymorphism enhanced the IRS1 association with IR, inhibited autophosphorylation of IR and IGF-1R, and reduced IRS1 phosphorylation level by 60% [47].
Likewise, the p.Arg1221Cys is located between two tyrosine phosphorylation sites that also serve as SHP2 binding sites: hTyr1179 (mTyr1172) and hTyr1229 (mTyr1222) [40, 48]. SHP2 is a member of the protein tyrosine phosphatase family encoded by the PTPN11 gene (NCBI Gene ID: 5781). It encompasses two SH2 domains, a catalytic phosphatase domain and the C-terminus. It is ubiquitously expressed in most tissues and is involved in many cellular signaling pathways [41]. Upon insulin stimulation and IRS1 association, SHP2 promotes IR endocytosis and the activation of the MAPK pathway that controls cell growth and proliferation [49]. Also, it differentially regulates the PI3K/AKT pathway by its tyrosine phosphatase activity, depending on cell types and upstream receptor kinases [50].
Mutations in the two tyrosine phosphorylation sites, hTyr1179 and hTyr1229, hindered the insulin-induced IRS1/SHP2 association [36]. Our variant is only two amino acids away from the widely conserved serine residues at position 1223 (mouse Ser1214). Phosphorylation of this serine residue weakens the IRS1/SHP2 association yet maintains the insulin-stimulated IRS1 tyrosine phosphorylation [51].
Both p.Pro948Leu and p.Arg1221Cys variants are globally rare (MAF are respectively 7.5 × 10-5 and 5.6 × 10-5), and they are never described together among the same individual (Table 2) even at heterozygote state. Intriguingly, the patient is homozygous for both variants’ minor alleles, suggesting their high frequencies in the Emirati population. This hypothesis has been rejected by analyzing a healthy Emirati cohort.
GDM and T2DM are similar in many aspects, such as their pathophysiological mechanisms, including increased insulin resistance, impaired insulin secretion, and β-cell impairment. Additionally, they share risk factors, such as obesity, age, history of abnormal glucose tolerance, family history of diabetes, and ethnicity [9, 52]. The mentioned risk factors interact with the environmental conditions making GDM a multifactorial metabolic disease. The genetic nature of GDM remains elusive, but much heritability evidence supports that there is a genetic background affecting its predisposition. Women with parental or grandparental diabetes tendencies, either maternal or paternal, exhibited a higher risk of GDM in many studies [53]. Genetic association studies have linked GDM to several genes involved in insulin resistance, insulin secretion, MODY, lipid and glucose metabolism, and other genes [13].
Given the crucial role of IRS1 in the insulin signaling pathway, many studies inspected its contribution to T2DM, especially the p.Gly972Arg variant in population-based investigations [43, 54]. IRS1 has also been investigated for GDM pathogenesis. Two meta-analysis studies by Zhang et al and Wu et al found the infamous p.Gly972Arg variant associated with increased GDM risk [12, 55, 56]. According to the DisGeNET database [57] and our Pubmed search, no other IRS1 variant was reported to be associated with GDM of different ethnicities. Here, despite the unavailability of the mother and sisters’ genotypes, we cannot neglect the effect of the two identified rare variants to affect insulin-insulin receptor signaling and the appearance of GDM. Considering the patient’s family history and her serological and genetic profiles, we excluded diabetic forms related to the MODY, type I diabetes, and LADA. The normal insulin level and high HOMA-IR reinforce the IRS-1 variant’s pathogenic effects. Consequently, new disease management has been proposed for the patient, marked by diet control and exercises with metformin, DPP4 inhibitors, pioglitazone, and SGL2i administration. Motivated by the insulin injection withdrawal and health improvement, the patient showed better compliance. Her HbA1c after 6 months of follow-up was < 7%, and she lost 5 kg of her body weight. Her blood pressure was better at 125/75 mm Hg with a resting heart rate of 87 bpm.
Genetic studies of multifactorial polygenic diseases such as GDM have inherent limitations, especially in a single case study where generalizability and small sample size are often questioned. However, they also present direct observations that can propose new possible risk variants. As stated above, the patient’s family members, unfortunately, did not consent to give samples to be sequenced. Thus, the co-segregation of the identified variants could not be perfectly assessed. The initial candidate gene approach was a source of potential bias towards the studied loci, which was circumvented by implementing the unbiased approach of CES. All these limitation factors should be considered when interpreting the results. In addition, a thorough functional investigation of these variants at the cellular level could undoubtedly bring more clarification regarding their individual and combined influence on protein expression, structure, and phosphorylation levels. It is also worthwhile to examine the potential downstream effect on the insulin receptor signaling pathway or association of IRS1 with SH2 domain-containing proteins.
Conclusion
CES and in silico analysis identified two rare IRS1 gene variants, p.Pro948Leu and p.Arg1221Cys, as possible risk candidates for GDM that require functional validation. Our findings suggest that some familial GDM forms could be analyzed as monogenic-like traits. The insulin resistance of GDM could be treated by lifestyle modifications, including a healthy diet, physical activity, losing excess weight, continuing oral antidiabetic medications, and stopping insulin injections.
| Supplementary Material | ▴Top |
Suppl 1. Sequence of primers used to amplify and sequence the exons of GCK (NG_008847.2) and HNF1A (NG_011731.2).
Suppl 2. Comparison of interactions between the wild and mutated protein residues in positions 948 and 1221 in wild type protein structure (A), variant p.Pro948Leu (B, C), and variant p.Arg1221Cys (D, E).
Suppl 3. Predicted stability changes (ΔΔG in kcal/mol) in IRS1 protein structure upon the selected mutations.
Acknowledgments
The authors would like to thank the patient and control subjects for their participation in this study. We would also like to show our gratitude to Mona Mahfood and Ruwaa Sandakli for their assistance with the control subjects.
Financial Disclosure
This research was funded by the Office of Vice Chancellor for Research and Graduate Studies from the University of Sharjah.
Conflict of Interest
Authors declare no conflict of interest in relation to this manuscript.
Informed Consent
Informed consent was obtained from the patient and control subjects included in this study.
Author Contributions
HK designed the study. AN recruited the patient and did the clinical examination. MB and HK performed the experiments, analyzed the data, and wrote the manuscript. UI conducted and analyzed the protein in silico analyses. All authors reviewed and approved the manuscript.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
| References | ▴Top |
- American Diabetes Association. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2021. Diabetes Care. 2021;44(Suppl 1):S15-S33.
doi pubmed - Peng TY, Ehrlich SF, Crites Y, Kitzmiller JL, Kuzniewicz MW, Hedderson MM, Ferrara A. Trends and racial and ethnic disparities in the prevalence of pregestational type 1 and type 2 diabetes in Northern California: 1996-2014. Am J Obstet Gynecol. 2017;216(2):177.e1-e8.
doi pubmed - Mission JF, Catov J, Deihl TE, Feghali M, Scifres C. Early pregnancy diabetes screening and diagnosis: prevalence, rates of abnormal test results, and associated factors. Obstet Gynecol. 2017;130(5):1136-1142.
doi pubmed - Chiefari E, Arcidiacono B, Foti D, Brunetti A. Gestational diabetes mellitus: an updated overview. J Endocrinol Invest. 2017;40(9):899-909.
doi pubmed - International Diabetes Federation. IDF Diabetes Atlas, 10th edn. Brussels, Belgium: 2021. Available at: https://www.diabetesatlas.org.
- Kampmann U, Knorr S, Fuglsang J, Ovesen P. Determinants of Maternal Insulin Resistance during Pregnancy: An Updated Overview. J Diabetes Res. 2019;2019:5320156.
doi pubmed - McIntyre HD, Catalano P, Zhang C, Desoye G, Mathiesen ER, Damm P. Gestational diabetes mellitus. Nature reviews Disease primers. 2019;5(1):1-19.
doi pubmed - Parsons JA, Brelje TC, Sorenson RL. Adaptation of islets of Langerhans to pregnancy: increased islet cell proliferation and insulin secretion correlates with the onset of placental lactogen secretion. Endocrinology. 1992;130(3):1459-1466.
doi pubmed - Buchanan TA, Xiang AH. Gestational diabetes mellitus. J Clin Invest. 2005;115(3):485-491.
doi - Zajdenverg L, Negrato CA. Gestational diabetes mellitus and type 2 diabetes: same disease in a different moment of life? Maybe not. Arch Endocrinol Metab. 2017;61(3):208-210.
doi pubmed - Lowe WL, Jr., Scholtens DM, Sandler V, Hayes MG. Genetics of Gestational Diabetes Mellitus and Maternal Metabolism. Curr Diab Rep. 2016;16(2):15.
doi pubmed - Zhang C, Bao W, Rong Y, Yang H, Bowers K, Yeung E, Kiely M. Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum Reprod Update. 2013;19(4):376-390.
doi pubmed - Robitaille J, Grant AM. The genetics of gestational diabetes mellitus: evidence for relationship with type 2 diabetes mellitus. Genet Med. 2008;10(4):240-250.
doi pubmed - https://primer3.ut.ee/.
- Moalla M, Safi W, Babiker Mansour M, Hadj Kacem M, Mahfood M, Abid M, Kammoun T, et al. Tunisian maturity-onset diabetes of the young: a short review and a new molecular and clinical investigation. Front Endocrinol (Lausanne). 2021;12:684018.
doi pubmed - https://sift.bii.a-star.edu.sg/.
- http://provean.jcvi.org/index.php.
- http://genetics.bwh.harvard.edu/pph2/.
- https://www.uniprot.org/.
- Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43(W1):W174-181.
doi pubmed - Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods. 2021;1(3):100014.
doi pubmed - Benkert P, Kunzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37(Web Server issue):W510-W514.
doi pubmed - Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J, Schwede T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics. 2020;36(6):1765-1771.
doi pubmed - Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407-W410.
doi pubmed - Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17(4):355-362.
doi pubmed - Chen Y, Lu H, Zhang N, Zhu Z, Wang S, Li M. PremPS: Predicting the impact of missense mutations on protein stability. PLoS Comput Biol. 2020;16(12):e1008543.
doi pubmed - Laimer J, Hofer H, Fritz M, Wegenkittl S, Lackner P. MAESTRO—multi agent stability prediction upon point mutations. BMC Bioinformatics. 2015;16:116.
doi pubmed - Sun X, Sui W, Wang X, Hou X, Ou M, Dai Y, Xiang Y. Whole-genome re-sequencing for the identification of high contribution susceptibility gene variants in patients with type 2 diabetes. Mol Med Rep. 2016;13(5):3735-3746.
doi pubmed - Zhou YJ, Yin RX, Hong SC, Yang Q, Cao XL, Chen WX. Association of the HNF1A polymorphisms and serum lipid traits, the risk of coronary artery disease and ischemic stroke. J Gene Med. 2017;19(1-2):e2941.
doi pubmed - Barzi SA, Ghaderian SM, Noormohammadi Z. A molecular case-control study of association of HNF1A gene polymorphisms (rs2259816 and rs7310409) with risk of coronary artery disease in Iranian patients. Hum Antibodies. 2017;25(1-2):65-70.
doi pubmed - Qi L, Parast L, Cai T, Powers C, Gervino EV, Hauser TH, Hu FB, et al. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J Am Coll Cardiol. 2011;58(25):2675-2682.
doi pubmed - Wakil SM, Muiya NP, Tahir AI, Al-Najai M, Baz B, Andres E, Mazhar N, et al. A new susceptibility locus for myocardial infarction, hypertension, type 2 diabetes mellitus, and dyslipidemia on chromosome 12q24. Dis Markers. 2014;2014:291419.
doi pubmed - Beysel S, Eyerci N, Pinarli FA, Kizilgul M, Ozcelik O, Caliskan M, Cakal E. HNF1A gene p.I27L is associated with early-onset, maturity-onset diabetes of the young-like diabetes in Turkey. BMC Endocr Disord. 2019;19(1):51.
doi pubmed - Zhu Y, Hu SC, Zheng PW, Jin MJ, Tang ML, Chen K, Wang JB. Association between CPR-related genetic variants and risk of ischemic stroke: a nested case-control study. Neurol Res. 2019;41(12):1090-1096.
doi pubmed - Wang XB, Han YD, Cui NH, Gao JJ, Yang J, Huang ZL, Zhu Q, et al. Associations of lipid levels susceptibility loci with coronary artery disease in Chinese population. Lipids Health Dis. 2015;14:80.
doi pubmed - Morjane I, Kefi R, Charoute H, Lakbakbi El Yaagoubi F, Hechmi M, Saile R, Abdelhak S, et al. Association study of HNF1A polymorphisms with metabolic syndrome in the Moroccan population. Diabetes Metab Syndr. 2017;11(Suppl 2):S853-S857.
doi pubmed - Lee YH, Gyu Song G. Genome-wide pathway analysis in pancreatic cancer. J BUON. 2015;20(6):1565-1575.
- Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55(10):2565-2582.
doi pubmed - Backer JM, Myers MG, Jr., Shoelson SE, Chin DJ, Sun XJ, Miralpeix M, Hu P, et al. Phosphatidylinositol 3'-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11(9):3469-3479.
doi pubmed - Grunberger G, Zick Y. Insulin signaling: from cultured cells to animal models. CRC Press. 2002.
doi - Sun XJ, Crimmins DL, Myers MG, Jr., Miralpeix M, White MF. Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol Cell Biol. 1993;13(12):7418-7428.
doi pubmed - Rocchi S, Tartare-Deckert S, Mothe I, Van Obberghen E. Identification by mutation of the tyrosine residues in the insulin receptor substrate-1 affecting association with the tyrosine phosphatase 2C and phosphatidylinositol 3-kinase. Endocrinology. 1995;136(12):5291-5297.
doi pubmed - Yamamoto-Honda R, Honda Z, Ueki K, Tobe K, Kaburagi Y, Takahashi Y, Tamemoto H, et al. Mutant of insulin receptor substrate-1 incapable of activating phosphatidylinositol 3-kinase did not mediate insulin-stimulated maturation of Xenopus laevis oocytes. J Biol Chem. 1996;271(45):28677-28681.
doi pubmed - Albegali AA, Shahzad M, Mahmood S, Ullah MI. Genetic association of insulin receptor substrate-1 (IRS-1, rs1801278) gene with insulin resistant of type 2 diabetes mellitus in a Pakistani population. Mol Biol Rep. 2019;46(6):6065-6070.
doi pubmed - Thangavelu M, Godla UR, Paul Solomon FD, Maddaly R. Single-nucleotide polymorphism of INS, INSR, IRS1, IRS2, PPAR-G and CAPN10 genes in the pathogenesis of polycystic ovary syndrome. J Genet. 2017;96(1):87-96.
doi pubmed - Alharbi KK, Khan IA, Abotalib Z, Al-Hakeem MM. Insulin receptor substrate-1 (IRS-1) Gly927Arg: correlation with gestational diabetes mellitus in Saudi women. Biomed Res Int. 2014;2014:146495.
doi pubmed - Xu Z, Yang H, Zhou X, Li J, Jiang L, Li D, Wu L, et al. Genetic variants in mTOR-pathway-related genes contribute to osteoarthritis susceptibility. Int Immunopharmacol. 2019;77:105960.
doi pubmed - McGettrick AJ, Feener EP, Kahn CR. Human insulin receptor substrate-1 (IRS-1) polymorphism G972R causes IRS-1 to associate with the insulin receptor and inhibit receptor autophosphorylation. J Biol Chem. 2005;280(8):6441-6446.
doi pubmed - Sugimoto S, Wandless TJ, Shoelson SE, Neel BG, Walsh CT. Activation of the SH2-containing protein tyrosine phosphatase, SH-PTP2, by phosphotyrosine-containing peptides derived from insulin receptor substrate-1. J Biol Chem. 1994;269(18):13614-13622.
doi pubmed - Choi E, Kikuchi S, Gao H, Brodzik K, Nassour I, Yopp A, Singal AG, et al. Mitotic regulators and the SHP2-MAPK pathway promote IR endocytosis and feedback regulation of insulin signaling. Nat Commun. 2019;10(1):1473.
doi pubmed - Myers MG, Jr., Mendez R, Shi P, Pierce JH, Rhoads R, White MF. The COOH-terminal tyrosine phosphorylation sites on IRS-1 bind SHP-2 and negatively regulate insulin signaling. J Biol Chem. 1998;273(41):26908-26914.
doi pubmed - Hausdorff SF, Bennett AM, Neel BG, Birnbaum MJ. Different signaling roles of SHPTP2 in insulin-induced GLUT1 expression and GLUT4 translocation. J Biol Chem. 1995;270(22):12965-12968.
doi pubmed - Luo M, Reyna S, Wang L, Yi Z, Carroll C, Dong LQ, Langlais P, et al. Identification of insulin receptor substrate 1 serine/threonine phosphorylation sites using mass spectrometry analysis: regulatory role of serine 1223. Endocrinology. 2005;146(10):4410-4416.
doi pubmed - Lewandowska M. Gestational Diabetes Mellitus (GDM) risk for declared family history of diabetes, in combination with BMI categories. Int J Environ Res Public Health. 2021;18(13):6936.
doi pubmed - Mahmutovic L, Bego T, Sterner M, Gremsperger G, Ahlqvist E, Velija Asimi Z, Prnjavorac B, et al. Association of IRS1 genetic variants with glucose control and insulin resistance in type 2 diabetic patients from Bosnia and Herzegovina. Drug Metab Pers Ther. 2019;34(1):20180031.
doi pubmed - Wu L, Cui L, Tam WH, Ma RC, Wang CC. Genetic variants associated with gestational diabetes mellitus: a meta-analysis and subgroup analysis. Sci Rep. 2016;6:30539.
doi pubmed - https://www.disgenet.org/.
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Endocrinology and Metabolism is published by Elmer Press Inc.
